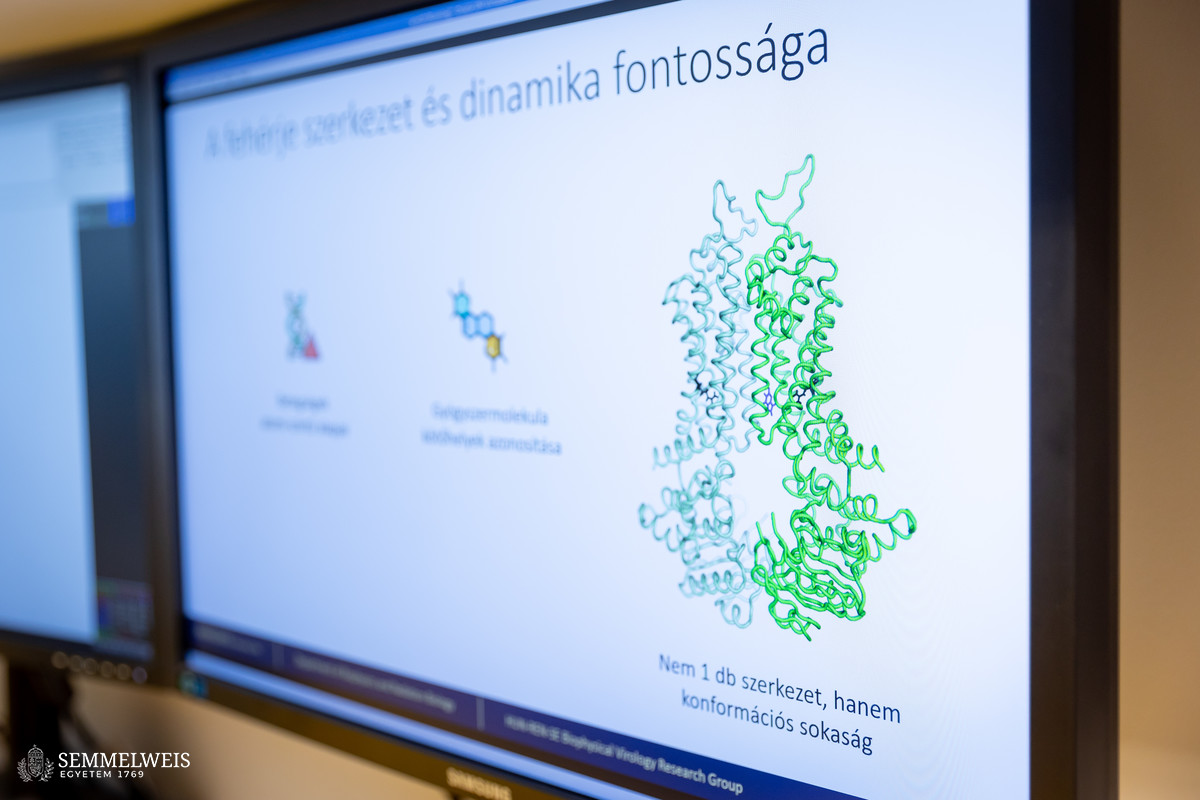
The 2024 Nobel Prize in Chemistry was awarded to three researchers: David Baker for his work on computational protein design, and Demis Hassabis and John M. Jumper for their work on protein structure prediction. David Baker is a Professor at the University of Washington (Seattle), who, together with his fellow researchers, has worked on predicting the spatial structure of proteins based on sequences. It is for this reason that he created the Rosetta software, which became widely used, primarily for protein design, but had limited applications for predicting protein structure. In contrast, Demis Hassabis and John M. Jumper, the lead researchers at Google DeepMind, created AlphaFold, a software tool that solved the problem of high-quality protein structure prediction and was also made freely available, revolutionizing many areas of 3D bioinformatics and ushering in a new era in life sciences, Dr. Tamás Hegedűs explained.
The 2024 Nobel Prize in Chemistry was awarded to three researchers: David Baker for his work on computational protein design, and Demis Hassabis and John M. Jumper for their work on protein structure prediction. David Baker is a Professor at the University of Washington (Seattle), who, together with his fellow researchers, has worked on predicting the spatial structure of proteins based on sequences. It is for this reason that he created the Rosetta software, which became widely used, primarily for protein design, but had limited applications for predicting protein structure. In contrast, Demis Hassabis and John M. Jumper, the lead researchers at Google DeepMind, created AlphaFold, a software tool that solved the problem of high-quality protein structure prediction and was also made freely available, revolutionizing many areas of 3D bioinformatics and ushering in a new era in life sciences, Dr. Tamás Hegedűs explained.
As the research professor at the Department of Biophysics and Radiation Biology pointed out, the exploration of the three-dimensional structure of proteins had long been a topic of interest for researchers. Many diseases can be traced back to mutations in the genes that code for proteins. Although knowledge of the structure of a protein is not always necessary for a cure, a precise understanding of it can be crucial for certain therapies or drug development, especially if the mutation affects the function or interactions of the protein.
According to Dr. Tamás Hegedűs, one of the most important technological advances of the past 15 years, which contributed to the breakthrough that was rewarded with last year’s Nobel Prize, was the dramatic development of graphics processing units (GPUs). These devices were originally designed to accelerate computer graphics, but due to their extraordinary parallel computing power they play a key role in scientific computing, such as in the field of artificial intelligence and deep learning algorithms. The Google DeepMind team developed AlphaFold, a program that can predict the spatial structure of proteins with extreme accuracy, by harnessing the power of GPUs and using neural networks.
Dr. Tamás Hegedűs and his colleagues published a highly cited article on the topic in 2022. In this paper, they investigated whether AlphaFold could also predict the structure of membrane proteins with sufficient accuracy, which is a particularly difficult task because the study of such proteins is challenging not only with experimental, but also with theoretical methods. Membrane proteins, however, play an important role in metabolic processes, signal transduction, and molecular transport, and more than half of the drugs currently available target membrane proteins, which is why it is so important to understand their structure. “We noticed that an error had been made in the experimental determination of the structure of one of the membrane proteins, so it had been incorrectly entered into the AlphaFold training set – yet the program was still able to predict the spatial structure of that protein correctly. In this way too, we demonstrated that AlphaFold could predict the structure of membrane proteins,” Dr. Tamás Hegedűs pointed out.
 The research professor at Semmelweis University co-authored a publication in 2022 with DeepMind researchers Demis Hassabis and John M. Jumper, who won the Nobel Prize last year. The paper is about the 3D-Beacon system, a new standard in structural bioinformatics. “As well as being able to predict the structure of proteins, AlphaFold has a huge database containing the structure of over 200 million proteins, and it is this database that has been made available to everyone. In addition, with the freely available AlphaFold server, almost anyone can now create structural models, so the number of such models has increased sharply,” Dr. Tamás Hegedűs explained. He noted that this increase in data had made it necessary to define standards that allow protein structural models and their data to be easily found, accessible, as well as usable and reusable by different programs; that is, to ensure FAIR (Findable, Accessible, Interoperable, Reusable) principles. This is the 3D-Beacon system, in the development of which Dr. Tamás Hegedűs was involved as a member of the management committee for 3D bioinformatics at ELIXIR, the European computing infrastructure consortium; while Demis Hassabis and John M. Jumper were involved in it as researchers at Google DeepMind.
The research professor at Semmelweis University co-authored a publication in 2022 with DeepMind researchers Demis Hassabis and John M. Jumper, who won the Nobel Prize last year. The paper is about the 3D-Beacon system, a new standard in structural bioinformatics. “As well as being able to predict the structure of proteins, AlphaFold has a huge database containing the structure of over 200 million proteins, and it is this database that has been made available to everyone. In addition, with the freely available AlphaFold server, almost anyone can now create structural models, so the number of such models has increased sharply,” Dr. Tamás Hegedűs explained. He noted that this increase in data had made it necessary to define standards that allow protein structural models and their data to be easily found, accessible, as well as usable and reusable by different programs; that is, to ensure FAIR (Findable, Accessible, Interoperable, Reusable) principles. This is the 3D-Beacon system, in the development of which Dr. Tamás Hegedűs was involved as a member of the management committee for 3D bioinformatics at ELIXIR, the European computing infrastructure consortium; while Demis Hassabis and John M. Jumper were involved in it as researchers at Google DeepMind.
 This new approach to 3D bioinformatics can contribute not only to more efficient and faster drug development, but also to the creation of better-targeted drugs and therapies, Dr. Tamás Hegedűs said. “Protein engineering can also be used to develop drugs that can be specifically tailored to a particular protein and its specific spatial structure. We can test thousands of designed proteins and millions of molecules to see which one contributes best to restoring the structure of the protein, but we can also test what concentration is most effective and whether the water solubility of the drug needs to be increased,” he noted. According to Dr. Tamás Hegedűs, the pace of progress in the discipline is so astonishing that even experts in the field are struggling to keep up. “Even in 2019, just before the release of the first version of AlphaFold, I was convinced that I would retire without it being possible to predict a high-quality structure from the amino acid sequence alone. In comparison, it actually happened within three years,” he pointed out.
This new approach to 3D bioinformatics can contribute not only to more efficient and faster drug development, but also to the creation of better-targeted drugs and therapies, Dr. Tamás Hegedűs said. “Protein engineering can also be used to develop drugs that can be specifically tailored to a particular protein and its specific spatial structure. We can test thousands of designed proteins and millions of molecules to see which one contributes best to restoring the structure of the protein, but we can also test what concentration is most effective and whether the water solubility of the drug needs to be increased,” he noted. According to Dr. Tamás Hegedűs, the pace of progress in the discipline is so astonishing that even experts in the field are struggling to keep up. “Even in 2019, just before the release of the first version of AlphaFold, I was convinced that I would retire without it being possible to predict a high-quality structure from the amino acid sequence alone. In comparison, it actually happened within three years,” he pointed out.
Ádám Szabó
Translation: Dr. Balázs Csizmadia
Photos by Boglárka Zellei – Semmelweis University