Learning opportunities
- Software development in C++ and Python
- Implementing machine learning algorithms (tensorlfow/keras, pytorch, scikit-learn)
- Data science fundamentals
- Biostatistics
- Network theory
Project leader: Olivér Balogh, MSc
Cellular processes are mediated by proteins and their interactions via physical contact, during which they form either stable long-term complexes or weak short-term ones. Given that proteins are not isolated inside the living cell, their functions are better understood if we consider the entire network of protein-protein interactions (PPIs), also called the protein interactome. Studying a system as a whole, rather than the sum of its parts, enables us to analyze emerging biological phenomena and to apply methods from other fields, such as network theory and artificial intelligence.
Visualization is a key part for the interpretation as well as presentation of results involving network analytical methods. Currently available visualizer frameworks offer several tools for this purpose, however, they often result in ‘hairball’ images, making network modules indistinguishable from the rest. In one of our projects, we are continuously developing a novel network visualization technique based on relative entropy minimization, available as the EntOptLayout plugin in the Cytoscape software. According to the principles formulated by Kovács et al. (Sci Rep, 2015), the algorithm considers the nodes as probability distributions and aims to select the optimal spatial representation with minimum module overlap and information loss through the minimization of the Kullback-Leibler divergence between the input and the output data. The EntOptLayout plugin enabled the visualization of large and often cluttered PPI networks, highlighting their emerging modules and major signaling complexes (Ágg et al., Bioinformatics 2019).
Artificial intelligence is not only a scientific field in itself, but also a tool with increasing presence in natural sciences, like medical imaging, drug-target discovery, protein structure prediction, pharmacovigilance signal detection, etc. In one of our projects, we adapt an artificial neural network, originally developed for computer vision tasks, for the problem of predicting the existence of previously unknown PPIs. Our approach relies only on the topological information contained in the given PPI network, without the need for molecular data, thus it performs link prediction in practice. The model utilizes a modified version of the generative adversarial network (GAN) architecture (Goodfellow et al. 2014), which implements a two-player mini-max game using two neural networks, called the generator and the discriminator. Our model (Balogh et al., BMC Bioinformatics, 2022) is modified in several aspects, like the use of conditional input (cGAN), convolutional layers and Wasserstein-distance based loss, then it is tasked to learn the increment of edges between differently connected subgraphs containing the same nodes. The results demonstrate that our model is an applicable and efficient method for PPI prediction, rivaling other non-AI based approaches (Wang et al. Nat. Commun. 2023).
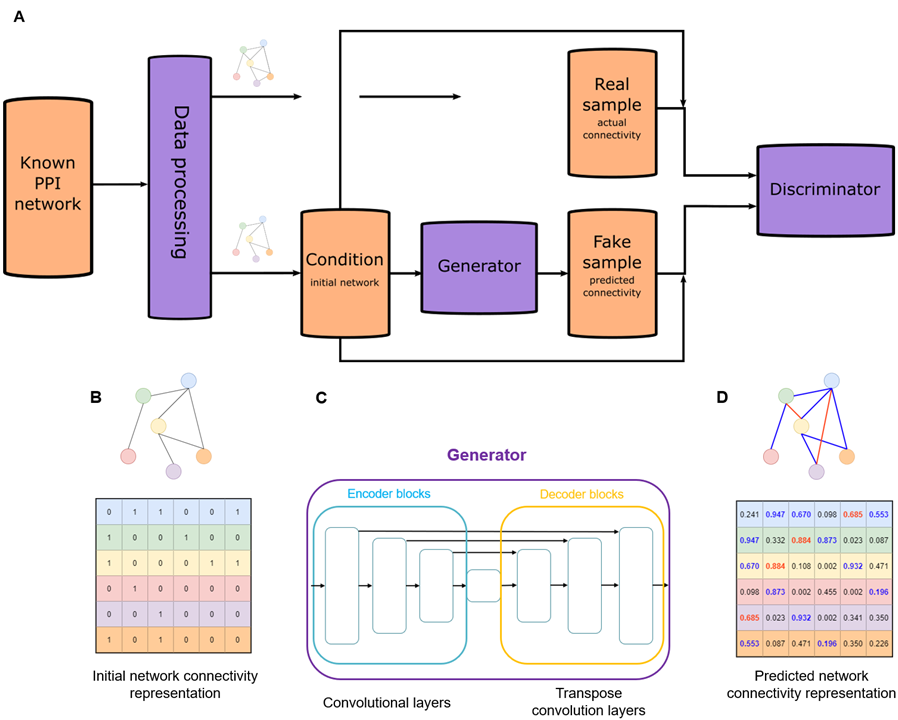
Figure 1.
Schematic diagram of the conditional generative adversarial network (cGAN) architecture that uses the representation of the initial protein-protein interaction (PPI) network connectivity as condition with no input noise in the generator, and pairs of condition and real or generated connectivity representations in the discriminator (A) and simplified visualization of the prediction process done by the generator (B-D). (Balogh et al., BMC Bioinformatics, 2022)
Publications in the field:
- Balogh, O. M., Benczik, B., Horváth, A., Pétervári, M., Csermely, P., Ferdinandy, P. & Ágg, B. Efficient link prediction in the protein–protein interaction network using topological information in a generative adversarial network machine learning model. BMC Bioinformatics 23, 1–19 (2022).
- Wang, X.-W., Madeddu, L., Spirohn, K., Martini, L., Fazzone, A., Becchetti, L., Wytock, T. P., Kovács, I. A., Balogh, O. M., Benczik, B., Pétervári, M., Ágg, B., Ferdinandy, P., Vulliard, L., Menche, J., Colonnese, S., Petti, M., Scarano, G., Cuomo, F., Hao, T., Laval, F., Willems, L., Twizere, J.-C., Vidal, M., Calderwood, M. A., Petrillo, E., Barabási, A.-L., Silverman, E. K., Loscalzo, J., Velardi, P. & Liu, Y.-Y. Assessment of community efforts to advance network-based prediction of protein–protein interactions. Nat. Commun. 2023 141 14, 1–14 (2023).
- Ágg, B., Császár, A., Szalay-Bekő, M., Veres, D. V, Mizsei, R., Ferdinandy, P., Csermely, P. & Kovács, I. A. The EntOptLayout Cytoscape plug-in for the efficient visualization of major protein complexes in protein–protein interaction and signalling networks. Bioinformatics 35, 4490 (2019).