The number of cancer patients is increasing worldwide (Thun, DeLancey, Center, Jemal, & Ward, 2010), and breast cancer is one of the most commonly diagnosed cancers, with 2.3 million new cases. (Sung et al., 2021). Tumours are genetically heterogeneous diseases characterised by a high variability of gene alterations. (Testa, Castelli, & Pelosi, 2020) However, “driver” genes that enable targeted therapy can be identified and are of major importance in disease development and therapy design (Shin, Bode, & Dong, 2017). In recent decades, a large number of new targeted therapeutics – effective for a particular gene alteration – have been approved (Bashraheel, Domling, & Goda, 2020). However, choosing the most appropriate therapy for a given patient remains a difficult task. This is what we are trying to find solutions to in our work.
1.1: Development and experimental validation of a precision oncology therapy algorithm
Project leaders: Orsolya Somogyi, András Makkos, MD, PhD 
Learning opportunities
– Cell culture methodologies, techniques
– Active lab work, from design to execution of experiments
– Light microscopy observation and imaging
– Fluorescence and luminescence analysis techniques
– Evaluation of experiments, application of statistical programs
– Abstracts, scientific presentation of results
– Making a presentation and presentation of results at a TDK conference
Project overview: Analysis of the individual genetic profile of a tumour can identify gene defects or genetic alterations that may contribute to the development or growth of the tumour. By combining molecular diagnostics with oncology software, it is possible to identify the potential molecular targets for tumor therapy and thus select the most appropriate targeted therapeutic treatment to target a specific tumor in a given patient.

The aim of our project, in collaboration with Oncompass Ltd, is to validate experimentally such a precision oncology therapy selection software on cell lines with known gene mutations.
.
1.2. ASelective CDK12 inhibition in combination with PARP inhibition sensitize PARP inhibitor resistant breast cancer cell lines
Project leaders: Orsolya Somogyi, András Makkos, MD, PhD
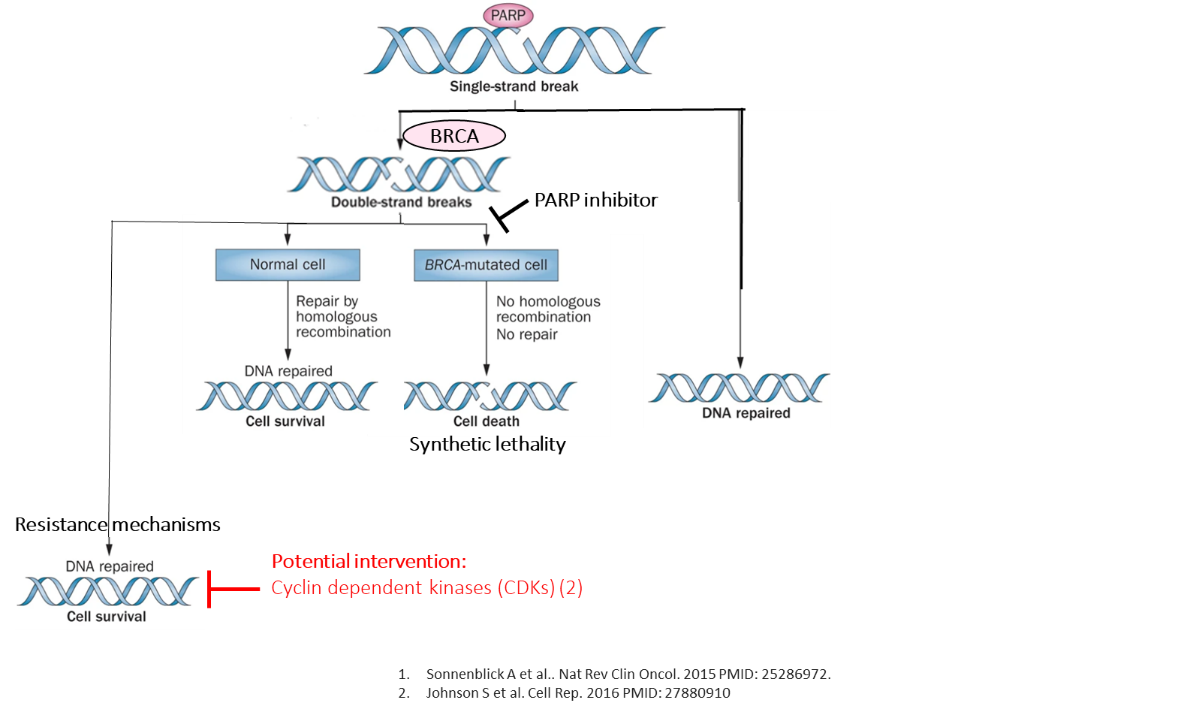
Project overview: DNA instability plays a role in the development of tumours. DNA damage can be repaired by homologous recombination mechanisms. Poly (ADP-ribose) polymerases (PARPs) and BRCA1/2 genes are key components of homologous recombination repair pathways. This forms the basis of PARP inhibitor therapy for patients with BRCA-mutant breast cancer. However, a proportion of tumours are resistant to PARP inhibitors. CDK12 gene expression may play a role in resistance to PARP inhibitors.
In this study, we aimed to investigate the role of CDK12 overexpression in the development of PARP inhibitor resistance. In this study, we aim to investigate the copy number and expression of CDK12 gene in breast cancer cell lines and the effect of CDK12 inhibitors on PARP inhibitor resistance.
Genetic instability is proportional to the number of mutations per unit time.