Nonsteroidal anti-inflammatory drugs (NSAIDs) are among the most widely used drugs worldwide. Their mucosal damaging effect in the stomach and duodenum is well-recognized and can be effectively prevented and treated by antisecretory drugs. However, NSAIDs also injure the lower gastrointestinal tract, and the development and introduction of new diagnostic tools (e.g. video capsule endoscopy) have revealed that small intestinal ulcers are much more common and serious than previously recognized (Bjarnason et al., 2018).
In the last two decades, much effort has been put into understanding the complex pathogenesis of NSAID enteropathy (Fig. 1), but it still remains insufficiently understood.
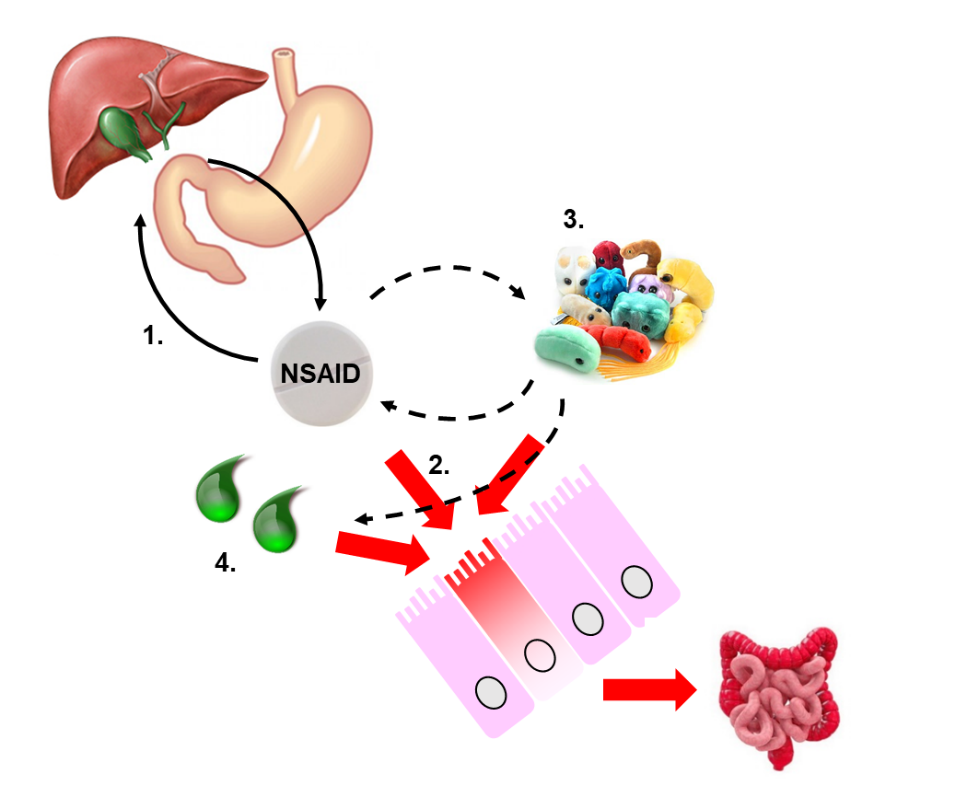
Our studies aim to identify novel mechanisms and targets in a rat model of NSAID enteropathy, which could potentially be useful for the design of new therapeutic interventions. After treating the animals with different NSAIDs, they are euthanized and their small intestines are removed. The severity of mucosal injury is evaluated macroscopically, histologically, and by various molecular biological techniques (Western blot, ELISA, qPCR, enzyme activity assays, etc.) (Fig. 2).

Considering the pivotal role of enteric bacteria and bile acids in the pathogenesis of enteropathy, we also aim to determine the composition of intestinal microflora in NSAID-treated animals (Fig. 3), as well as the concentrations of primary and secondary bile acids.

Acquirable skills: drug treatment of animals, macroscopic evaluation of enteropathy, histology, Western blot, ELISA
Tutor: Dr. Zoltán Zádori, associate professor
Our previous publications related to this project:
- Zádori ZS, Király K, Al-Khrasani M, Gyires K. Interactions between NSAIDs, opioids and the gut microbiota – Future perspectives in the management of inflammation and pain. Pharmacol Ther. 2023 241:108327.
- Hutka B, Lázár B, Tóth AS, Ágg B, László SB, Makra N, Ligeti B, Scheich B, Király K, Al-Khrasani M, Szabó D, Ferdinandy P, Gyires K, Zádori ZS. The Nonsteroidal Anti-Inflammatory Drug Ketorolac Alters the Small Intestinal Microbiota and Bile Acids Without Inducing Intestinal Damage or Delaying Peristalsis in the Rat.Front Pharmacol. 2021 12:664177.
- Lázár B, László SB, Hutka B, Tóth AS, Mohammadzadeh A, Berekméri E, Ágg B, Balogh M, Sajtos V, Király K, Al-Khrasani M, Földes A, Varga G, Makra N, Ostorházi E, Szabó D, Ligeti B, Kemény Á, Helyes Z, Ferdinandy P, Gyires K, Zádori ZS. A comprehensive time course and correlation analysis of indomethacin-induced inflammation, bile acid alterations and dysbiosis in the rat small intestine.Biochem Pharmacol. 2021 190:114590.
- Lázár B, Brenner GB, Makkos A, Balogh M, László SB, Al-Khrasani M, Hutka B, Bató E, Ostorházi E, Juhász J, Kemény Á, László T, Tiszlavicz L, Bihari Z, Giricz Z, Szabó D, Helyes Z, Ferdinandy P, Gyires K, Zádori ZS. Lack of Small Intestinal Dysbiosis Following Long-Term Selective Inhibition of Cyclooxygenase-2 by Rofecoxib in the Rat. Cells. 2019 8(3):251.