Kutatócsoport neve: TRAJECTOME Kutatócsoport
Projekt címe: Depresszió alcsoportok azonosítása időbeli multimorbiditás hálózatok alapján: a betegségutak kvantitatív vizsgálata és lehetséges gyógyszercélpontok felkutatása
Projekt rövidítése: TRAJECTOME
Koordinátor:
Semmelweis Egyetem, Gyógyszerhatástani Intézet
Dr. Juhász Gabriella, PhD, egyetemi docens
E-mail: juhasz.gabriella@semmelweis.hu
Tel: +36 1 459 1500
A projekt partnerei:
- Abiomics Europe Ltd., Budapest, Hungary, Dr. Antal Péter
- The Department of Public Health Solutions, Finnish Institute for Health and Welfare, Helsinki, Finland, Dr. Mikko Kuokkanen
- Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS) – Hospital Clinic de Barcelona, Barcelona, Spain, Dr. Josep Roca
- Department of Psychiatry, University Medicine Greifswald (UMG), Greifswald, Mecklenburg-West Pomerania, Germany, Dr. rer. med. Sandra Van der Auwera-Palitschka
Projekt megvalósításának kezdete: 2020.05.01.
Projekt megvalósításának befejezése: 2023.04.30.
Link: https://semmelweis.hu/gyogyszerhatastan/TRAJECTOME/
 A projekt kiírója: ERA PerMed Joint Translational Call 2019, “Personalised Medicine: Multidisciplinary Research Towards Implementation” (pályázat száma: ERAPERMED2019-108)
A projekt kiírója: ERA PerMed Joint Translational Call 2019, “Personalised Medicine: Multidisciplinary Research Towards Implementation” (pályázat száma: ERAPERMED2019-108)
További információk a projekt kiírójáról: https://erapermed.isciii.es/
Támogató: Nemzeti Kutatási, Fejlesztési és Innovációs Hivatal (projekt/ szerződés száma: 2019-2.1.7-ERA-NET-2020-00005)
Támogatás összege: konzorcium szinten 1,115M EUR, amiből az egyetem részére 150 000 EUR
A kutatásról
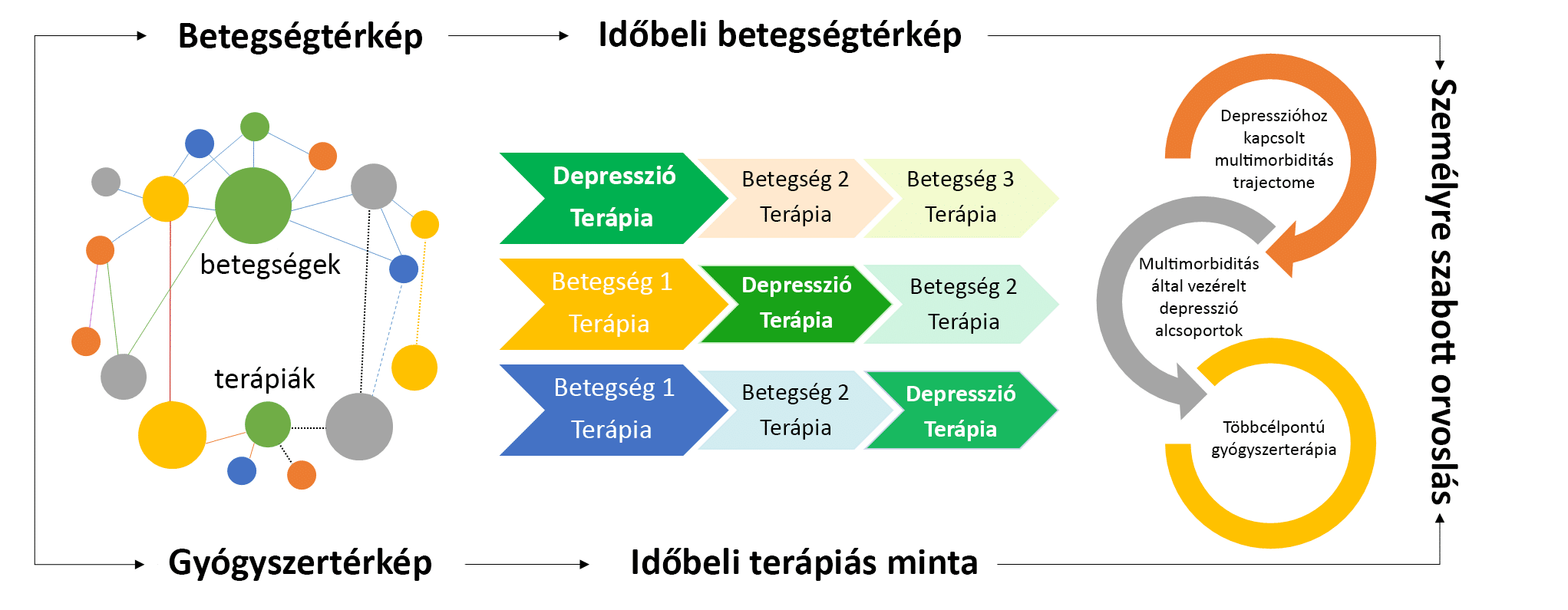
A depresszió az egyik leggyakoribb mentális betegség, mely általában krónikus jellegű, az élet folyamán ismételten visszatérő állapotrosszabbodásokkal jellemezhető, ezzel jelentős szenvedést okozva a betegeknek. Korábbi kutatások kimutatták, hogy a depressziót átélt személyekben gyakrabban alakulnak ki más lelki és testi betegségek is, úgynevezett multimorbid állapotok. Ennek oka valószínűleg a különböző betegségek között fennálló közös biológiai és környezeti rizikófaktorokra vezethető vissza.
A TRAJECTOME projekt célja, hogy megvizsgálja, milyen időbeli sorrendben alakulnak ki a depresszióhoz kapcsolódó betegségek, majd meghatározza a leggyakrabban előforduló betegség-sorrendeket, más néven betegségutakat.
A különböző betegségutak azonosítása lehetővé teszi, hogy a hasonló betegségekkel rendelkező depressziós személyeket alcsoportokba soroljuk, majd meghatározzuk a kiválasztott alcsoportra jellemző specifikus biológiai, társadalmi-gazdasági és életmódbeli rizikófaktorokat, melyek hozzájárulnak a depresszió tüneteinek kialakulásához. Ezeknek a folyamatoknak a megértésével a TRAJECTOME projekt elősegíti a depresszió kockázatának pontosabb becslését és hatékonyabb betegség megelőzési stratégiák kidolgozását. Ezen kívül új irányvonalat mutathat a gyógyszerkutatás számára is, mely hozzájárul a depresszióval összefüggő betegségek személyre szabott kezelésének kidolgozásához.