To be able to discover novel drug targets we aim to develop software based on network theoretic approaches that are capable to identify mediators and pathways involved in the pathomechanism of various cardiovascular diseases by the analysis of datasets assessed with high throughput molecular biological techniques (e.g. microarray, RNA-seq) of samples gathered from small and large animal models of the studied cardiovascular disorders. Our main focus is the analysis of the transcriptomics datasets, including microRNA fingerprints.
Learning opportunities:
- Bioinformatics methodologies
- Analysis of raw RNA sequencing data
- Biostatistics
- Writing R and UNIX/Linux shell scripts
- Network theory
1.1. MicroRNA-target interaction network analysis
Project leader: Bettina Benczik, MSc
We aim to develop software based on network theoretic approaches that are capable of identifying mediators and pathways involved in the pathomechanism of various cardiovascular diseases by the analysis of datasets assessed with high throughput molecular biological techniques (e.g. microarray, RNA-seq) of samples gathered from small and large animal models of the studied cardiovascular disorders. Our main focus is the analysis of the transcriptomics datasets, including microRNA fingerprints.
MicroRNAs (miRNAs) are short (18-24 nt) RNA sequences that could decrease the expression of various target genes both on the messenger RNA (mRNA) and the protein level. As miRNAs could have multiple hundreds of targets, and also one mRNA could be regulated by dozens of miRNAs the post-transcriptional regulatory network of miRNAs are extremely complex (Figure 1). Therefore, to understand and predict the effect of changes in the miRNA expression profiles measured by the so called omics (global, high throughput) methodologies, sophisticated bioinformatics and network theoretic approaches are necessary.
In this project we aim to develop, implement and utilize such network theoretic models to efficiently search for drug target candidates in various cardiovascular diseases.
We also aim to make it possible for researchers without previous bioinformatics experiences to analyze miRNA expression datasets with user friendly tools. For this purpose our team in collaboration with the Pharmahungary Group has developed the miRNAtarget.com web application, which could be used to predict common targets of differentially expressed miRNAs.
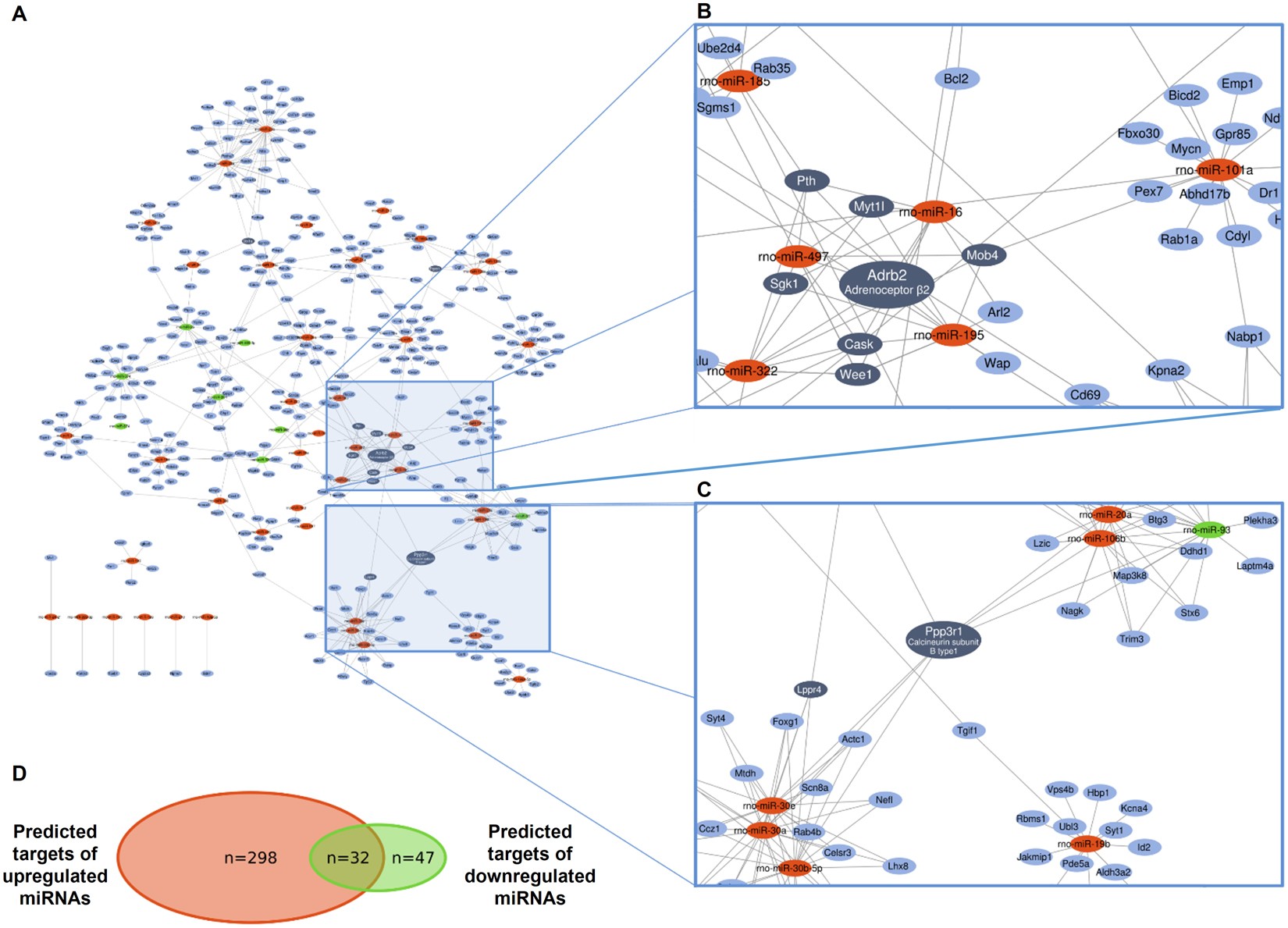
Figure 1. An example of miRNA-target interaction network. Downregulated, upregulated miRNAs and mRNAs are indicated in green, red and blue, respectively. Dark blue represents mRNAs with at least 4 target interactions. (Ágg et al, Sci Rep 2018)
1.2. Bioinformatics analysis of RNA sequencing datasets
Project leader: Dr. Barnabás Váradi, MD
The starting point for an unbiased, hypothesis-free analysis of various diseases is the assessment of global transcriptomics profiles. Nowadays the most well-established and most cost-effective approach for transcriptomics measurements is RNA sequencing. RNA sequencing produces huge raw datasets, which need to be analyzed by various bioinformatics methods.
Our group is continuously developing, adapting and utilizing bioinformatics and statistical methodologies to be able to better understand RNA sequencing results.
Our aims are to develop tools to be able:
- to integrate and analyze data from RNA sequencing and other sources
- to facilitate data interpretation by dimensionality reduction and customized distance metrics (Figure 2.)
- to visualize results to facilitate hypothesis generation
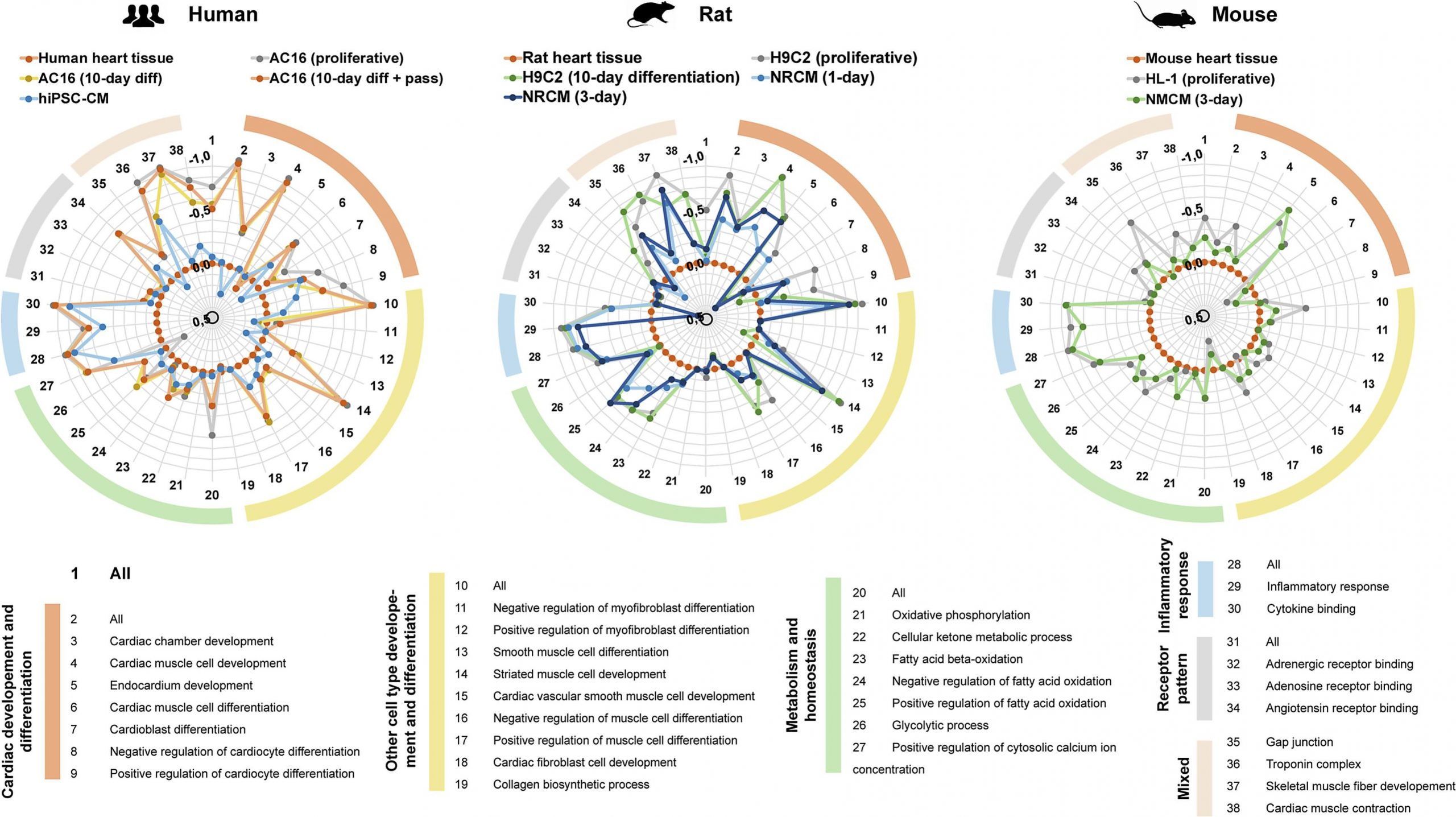
Figure 2. Radar plot with the dissimilarity of the Gene Ontology term-based functional expression vectors of cardiac cell lines and primary cultures compared to the corresponding reference tissue in human, rat and mouse species (Onódi et al, J Mol Cell Cardiol 2022)
Publications in the field:
- Varga ZV, Ágg B, Ferdinandy P. miR-125b is a protectomiR: A rising star for acute cardioprotection. J Mol Cell Cardiol. 2018 Feb;115:51-53.
- Schulz R, Ágg B, Ferdinandy P. Survival pathways in cardiac conditioning: individual data vs. meta-analyses. What do we learn? Basic Res Cardiol. 2017 Nov 21;113(1):4.
- Perrino C, Barabási AL, Condorelli G, Davidson SM, De Windt L, Dimmeler S, Engel FB, Hausenloy DJ, Hill JA, Van Laake LW, Lecour S, Leor J, Madonna R, Mayr M, Prunier F, Sluijter JPG, Schulz R, Thum T, Ytrehus K, Ferdinandy P. Epigenomic and transcriptomic approaches in the post-genomic era: path to novel targets for diagnosis and therapy of the ischaemic heart? Position Paper of the European Society of Cardiology Working Group on Cellular Biology of the Heart. Cardiovasc Res. 2017 Jun 1;113(7):725-736.
- Ágg B, Baranyai T, Makkos A, Vető B, Faragó N, Zvara Á, Giricz Z, Veres DV, Csermely P, Arányi T, Puskás LG, Varga ZV, Ferdinandy P. MicroRNA interactome analysis predicts post-transcriptional regulation of ADRB2 and PPP3R1 in the hypercholesterolemic myocardium. Sci Rep. 2018 Jul 4;8(1):10134.
- Bencsik P, Kiss K, Ágg B, Baán JA, Ágoston G, Varga A, Gömöri K, Mendler L, Faragó N, Zvara Á, Sántha P, Puskás LG, Jancsó G, Ferdinandy P. Sensory Neuropathy Affects Cardiac miRNA Expression Network Targeting IGF-1, SLC2a-12, EIF-4e, and ULK-2 mRNAs. Int J Mol Sci. 2019 Feb 25;20(4):991.
- Sághy É, Vörös I, Ágg B, Kiss B, Koncsos G, Varga ZV, Görbe A, Giricz Z, Schulz R, Ferdinandy P. Cardiac miRNA Expression and their mRNA Targets in a Rat Model of Prediabetes. Int J Mol Sci. 2020 Mar 20;21(6):2128.
- Schreckenberg R, Klein J, Kutsche HS, Schulz R, Gömöri K, Bencsik P, Benczik B, Ágg B, Sághy É, Ferdinandy P, Schlüter KD. Ischaemic post-conditioning in rats: Responder and non-responder differ in transcriptome of mitochondrial proteins. J Cell Mol Med. 2020 May;24(10):5528-5541.
- Róka B, Tod P, Kaucsár T, Bukosza ÉN, Vörös I, Varga ZV, Petrovich B, Ágg B, Ferdinandy P, Szénási G, Hamar P. Delayed Contralateral Nephrectomy Halted Post-Ischemic Renal Fibrosis Progression and Inhibited the Ischemia-Induced Fibromir Upregulation in Mice. Biomedicines. 2021 Jul 14;9(7):815.
- Makkos A, Ágg B, Petrovich B, Varga ZV, Görbe A, Ferdinandy P. Systematic review and network analysis of microRNAs involved in cardioprotection against myocardial ischemia/reperfusion injury and infarction: Involvement of redox signalling. Free Radic Biol Med. 2021 Aug 20;172:237-251.
- Makkos A, Ágg B, Varga ZV, Giricz Z, Gyöngyösi M, Lukovic D, Schulz R, Barteková M, Görbe A, Ferdinandy P. Molecular Network Approach Reveals Rictor as a Central Target of Cardiac ProtectomiRs. Int J Mol Sci. 2021 Sep 2;22(17):9539.
- Vörös I, Sághy É, Pohóczky K, Makkos A, Onódi Z, Brenner GB, Baranyai T, Ágg B, Váradi B, Kemény Á, Leszek P, Görbe A, Varga ZV, Giricz Z, Schulz R, Helyes Z, Ferdinandy P. Somatostatin and Its Receptors in Myocardial Ischemia/Reperfusion Injury and Cardioprotection. Front Pharmacol. 2021 Nov 5;12:663655.
- Gáspár R, Gömöri K, Kiss B, Szántai Á, Pálóczi J, Varga ZV, Pipis J, Váradi B, Ágg B, Csont T, Ferdinandy P, Barteková M, Görbe A. Decorin Protects Cardiac Myocytes against Simulated Ischemia/Reperfusion Injury. Molecules. 2020 Jul 28;25(15):3426.
- Onódi Z, Visnovitz T, Kiss B, Hambalkó S, Koncz A, Ágg B, Váradi B, Tóth VÉ, Nagy RN, Gergely TG, Gergő D, Makkos A, Pelyhe C, Varga N, Reé D, Apáti Á, Leszek P, Kovács T, Nagy N, Ferdinandy P, Buzás EI, Görbe A, Giricz Z, Varga ZV. Systematic transcriptomic and phenotypic characterization of human and murine cardiac myocyte cell lines and primary cardiomyocytes reveals serious limitations and low resemblances to adult cardiac phenotype. J Mol Cell Cardiol. 2022 Apr;165:19-30.
- Aczél T, Benczik B, Ágg B, Körtési T, Urbán P, Bauer W, Gyenesei A, Tuka B, Tajti J, Ferdinandy P, Vécsei L, Bölcskei K, Kun J, Helyes Z. Disease- and headache-specific microRNA signatures and their predicted mRNA targets in peripheral blood mononuclear cells in migraineurs: role of inflammatory signalling and oxidative stress. J Headache Pain. 2022 Sep 2;23(1):113.